Connect code and reports with
Typical guidelines for keeping a notebook of wet-lab work¶
- Record everything you do in the lab, even if you are following a published procedure.
- If you make a mistake, put a line through the mistake and write the new information next to it.
- Use a ball point pen so that marks will not smear nor will they be erasable.
- Use a bound notebook so that tear-out would be visible.
- When you finish a page, put a corner-to corner line through any blank parts that could still be used for data entry.
- All pages must be pre-numbered.
- Write a title for each and every new set of entries.
- It is critical that you enter all procedures and data directly into your notebook in a timely manner.
- Properly introduce and summarize each experiment.
- The investigator and supervisor must sign each page.
etc...
Typical guidelines for keeping a notebook of dry-lab work¶

Literate programming
Instead of imagining that our main task is to instruct a computer what to do, let us concentrate rather on explaining to human beings what we want a computer to do. - Donald Knuth (1984)
Literate computing
A literate computing environment is one that allows users not only to execute commands interactively, but also to store in a literate document the results of these commands along with figures and free-form text. - Millman KJ and Perez F (2014)
Wolfram Mathematica notebook (1987)

The Jupyter notebook¶
The Jupyter Notebook is a web application for interactive data science and scientific computing.
In-browser editing for code, with automatic syntax highlighting, indentation, and tab completion/introspection.
Document your work in Markdown
Penguin data analysis¶
Here we will investigate the Penguin dataset.
The species included in this set are:
- Adelie
- Chinstrap
- Gentoo
Execute code directly from the browser, with results attached to the code which generated them
data = sns.load_dataset("penguins")
data.groupby("species").mean()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| species | ||||
| Adelie | 38.791391 | 18.346358 | 189.953642 | 3700.662252 |
| Chinstrap | 48.833824 | 18.420588 | 195.823529 | 3733.088235 |
| Gentoo | 47.504878 | 14.982114 | 217.186992 | 5076.016260 |
Generate plots directly in the browser and/or save to file.
ax = sns.pairplot(data, hue="species", height=1,
plot_kws=dict(s=20, linewidth=0.5),
diag_kws=dict(linewidth=0.5))
Mix and match languages in addition to python (e.g. R, bash, ruby)
%%R
x <- 1:12
sample(x, replace = TRUE)
[1] 2 1 9 12 6 3 7 4 2 6 6 3
%%bash
uname -v
Darwin Kernel Version 19.6.0: Tue Oct 12 18:34:05 PDT 2021; root:xnu-6153.141.43~1/RELEASE_X86_64
Create interactive widgets
def f(palette, x, y):
plt.figure(1, figsize=(3,3))
ax = sns.scatterplot(data=data, x=x, y=y, hue="species", palette=palette)
ax.legend(bbox_to_anchor=(1,1))
_ = interact(f, palette=["Set1","Set2","Dark2","Paired"],
y=["bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g"],
x=["bill_depth_mm", "bill_length_mm", "flipper_length_mm", "body_mass_g"])
Notebook basics¶
- Runs as a local web server
- Load/save/manage notebooks from the menu

The notebook itself is a JSON file
!head -20 jupyter.ipynb
{
"cells": [
{
"cell_type": "code",
"execution_count": 2,
"metadata": {
"slideshow": {
"slide_type": "skip"
}
},
"outputs": [],
"source": [
"import seaborn as sns\n",
"import pandas as pd\n",
"import matplotlib.pyplot as plt\n",
"from ipywidgets import interact\n",
"%matplotlib inline\n",
"%config InlineBackend.figure_format = 'svg'\n",
"plt.style.use('seaborn-talk')"
]
Sharing is caring¶
- Put the notebook on GitHub/Bitbucket and it will be rendered there...
- ... or export to one of many different formats, e.g. HTML, PDF, code, slides etc. (this presentation is a Jupyter notebook)
Or paste a link to any Jupyter notebook at nbviewer.jupyter.org and it will be rendered for you.
%%html
<!-- MRSA Notebook that you'll work on in the tutorial -->
<!-- https://github.com/NBISweden/workshop-reproducible-research/blob/main/tutorials/jupyter/supplementary_material.ipynb -->
<iframe src="https://nbviewer.jupyter.org/" height="800" width="800"></iframe>
Or generate interactive notebooks using Binder
%%HTML
<iframe src="https://mybinder.org" height="800" width="800"></iframe>
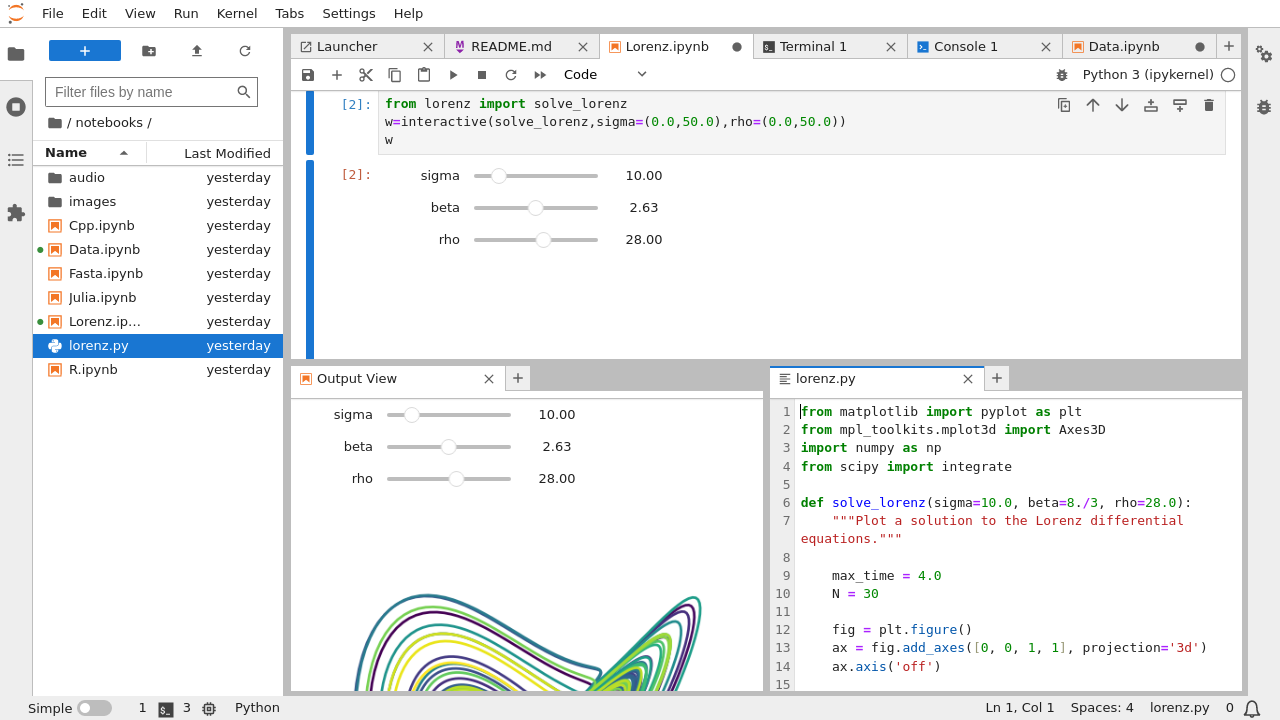
- full-fledged IDE, similar to e.g. Rstudio.
- Tab views, Code consoles, Show output in a separate tab, Live rendering of edits
conda install –c conda-forge jupyterlab
lets you build an online book using a collection of Jupyter Notebooks and Markdown files
- Interactivity
- Citations
- Build and host it online with GitHub/GitHub Pages...
- or locally on your own laptop