|
Introduction to Oxford Nanopore Technology data analyses for metaviromicsThis course offers an introduction to ONT data analysis with a focus in virus reconstruction. It includes 4 issues: basecalling, reads quality control, taxonomic assignation (diamond, kaiju and kraken2) and assemblies (incuding polishing/correction). |
Prerequisites
Linux and knowledge of NGS formats
Program
- Introduction to the principles of the technology (kits, flowcell)
- Principles of basecalling
- QC and data cleaning
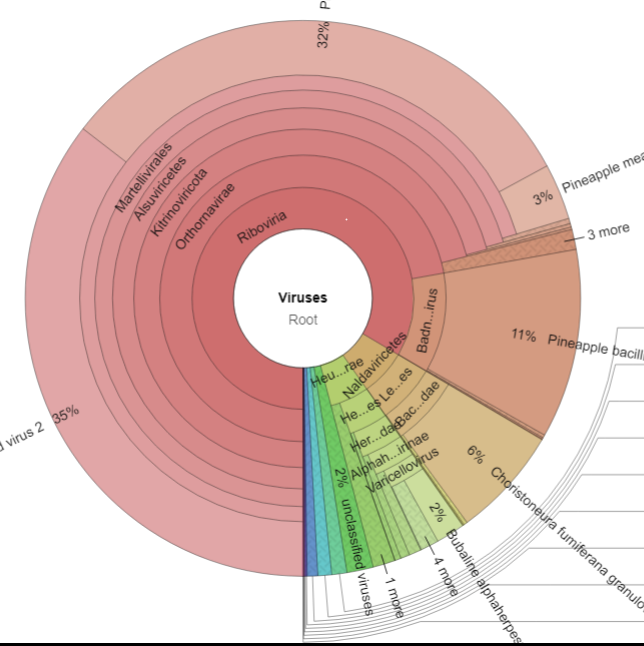
- taxonomic assignation with diamond and kaiju and vizualisation
- Handling mapping and assembly tools suitable for ONT data
- How to assemble a metagenome
Learning objectives
- Understanding limits and advantages of ONT technology
- Manipulating ONT data on a virtual machine on jupyter environment
- Handling metagenomics tools to be able to analyse your own data
Instructors
- Julie Orjuela (JO) - julie.orjuela@ird.fr UMR DIADE IRD
- Denis Filloux (DF) - denis.filloux@cirad.fr UMR PHIM IRD
- Aurore Comte (AC) - aurore.comte@ird.fr UMR PHIM CIRAD
Trainings
| Date | Location | Topics | Participants | Instructors | Links | Units |
| 2022 | Burkina Faso | Introduction to ONT data analyses for viruses | 10 | JO, DF, AC | UMR DIADE, PHIM |
