|
Introduction to Structural variant detection analysesThis course offers an introduction to SNP and SV detection data analysis by using advantages of short and long sequencing technologies. |
Prerequisites
Linux and knowledge of NGS formats Patience and perseverance
Program
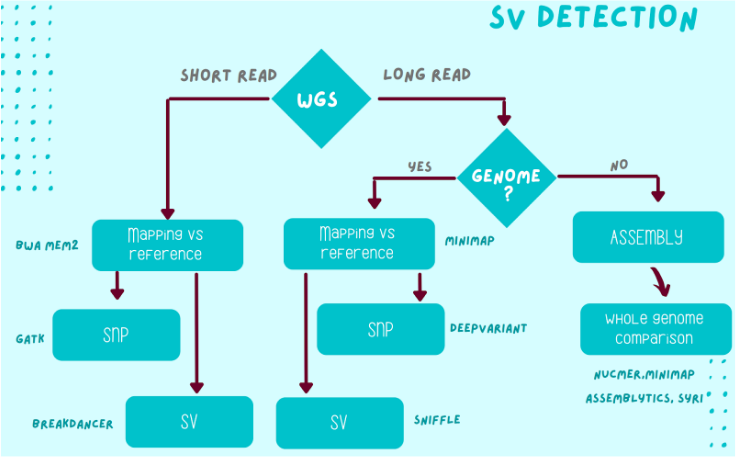
- Handling mapping tools suitable for ILLUMINA and ONT data (bwa, minimap2)
- SNP detection from mapping of short reads against a reference genome: SNP calling, filters and SNP annotation. Examples of possible studies based on SNP arrays
- Detecting Structural Variations (SV) in short and long reads (breakdancer, sniffle)
- SV detection from genome assembly and comparison (minimap2, nucmer, assemblytics, siry)
Learning objectives
- Understanding limits and advantages of sequencing technology for SV detection
- Manipulating short and long reads data on a virtual machine on jupyter environment
- Handling mapping, SNP calling and SV detection tools and be able to analyse your own data
- Detecting structural variations comparing assemblies.
Instructors
- Christine Tranchant (CT) - christine.tranchant@ird.fr
- Francois Sabot (FS) - francois.sabot@ird.fr
- Julie Orjuela (JO) - julie.orjuela@ird.fr
- Alexis Dereeper (AD) - alexis.dereeper@ird.fr
Trainings
| Date | Location | Topics | Participants | Instructors | Links | Units |
| 2022 | Ouagadougour, Burkina Faso | Introduction to SV detecton analyses | 12 | CT, JO, AD | UMR DIADE, PHIM | |
| 2022 | Montpellier, France | Introduction to SV detecton analyses | 15 | CT, FS, JO, AD | UMR DIADE, PHIM |
