| Description | Hands On Lab Exercises for Artemis |
|---|---|
| Related-course materials | Structural annotation with MAKER Structural annotation with EGN-EP |
| Authors | Stéphanie Bocs (stephanie.sidibe-bocs@cirad.fr) Lucile Soler (lucile.soler@nbis.se) Jacques Dainat (jacques.dainat@nbis.se) |
| Creation Date | 26/09/2018 |
| Last Modified Date | 27/09/2018 |
Summary
- Goal of the exercise
- Install & configure Artemis
- Get annotation and evidence gff3 files
- Launch Artemis & Load data
- Some Artemis functionalities
- Annotate AT4G32500
- Annotate AT4G32510
Goal of the exercise
- General: Become familiar with the Artemis annotation tool with regard to manual gene curation.
- Specific: Annotate two genes (AT4G32500 and AT4G32510) of the whole Arabidopsis chromosome 4 from the MAKER gene prediction with functional annotation
Install & configure artemis :
Download Artemis from github.io (e.g. v17 20180928).
Increase the memory allocated to Artemis following the FAQ recommendations For instance -Xmx8g (Java max heap size) instead of -Xmx2g in Artemis.cfg if you have 16Gb of RAM on your personal computer
Get annotation and evidence gff3 files :
To facilitate manual annotation, match_part must be linked in Artemis, so gff files need to be reformatted. For your information, from the Eugene-EP IFB appliance:
/root/work_dir/0001/Chr4/
awk 'BEGIN{g=0}{if($3 ~ /EST_match/){g++; print $1"\t"$2"\tgene\t"$4"\t"$5"\t"$6"\t"$7"\t"$8"\tID="g; print $1"\t"$2"\tmRNA\t"$4"\t"$5"\t"$6"\t"$7"\t"$8"\t"$9";Parent="g}else{if($3 ~ /match_part/){print $0}else{print $0}}}' Chr4.est1.gff3 |sed 's/match_part/exon/' > Chr4.est1_gene.gff3Reformatted data are centralised on the Slovenian VM, for your information MAKER formatting
awk 'BEGIN{g=0}{if($3 ~ /protein_match/){g++; print $1"\t"$2"\tgene\t"$4"\t"$5"\t"$6"\t"$7"\t"$8"\tID="g; print $1"\t"$2"\tmRNA\t"$4"\t"$5"\t"$6"\t"$7"\t"$8"\t"$9";Parent="g}else{if($3 ~ /match_part/){print $0}else{print $0}}}' protein_gff\:protein2genome.gff |sed 's/match_part/CDS/' > protein2genome_gene.gff
awk 'BEGIN{g=0}{if($3 ~ /^match$/){g++; print $1"\t"$2"\tgene\t"$4"\t"$5"\t"$6"\t"$7"\t"$8"\tID="g; print $1"\t"$2"\tmRNA\t"$4"\t"$5"\t"$6"\t"$7"\t"$8"\t"$9";Parent="g}else{if($3 ~ /match_part/){print $0}else{print $0}}}' augustus_masked.gff |sed 's/match_part/exon/' > augustus_masked_gene.gffTransfer the reformatted annotation files from the Slovenian VM to your personal computer
scp -r -P 65034 gaas23@terminal.mf.uni-lj.si:/home/data/byod/Annotation/ARATH/ARATH04_MAKER/ .
scp -r -P 65034 gaas23@terminal.mf.uni-lj.si:/home/data/byod/Annotation/ARATH/ARATH04_EGN .Launch artemis & Load data :
-
File/Open Tous les fichiers (All files) Chr4
-
File/Read An Entry
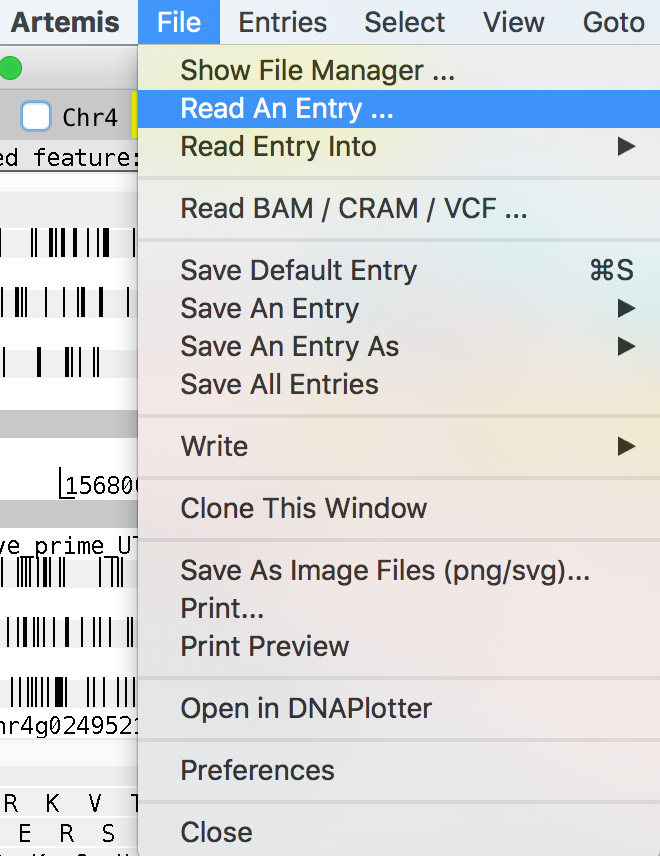
maker_abinitio_functional.gff
Chr4_egnep.gff3
Chr4.est1_gene.gff3
protein2genome_gene.gff
augustus_masked.gff
Some Artemis functionalities :
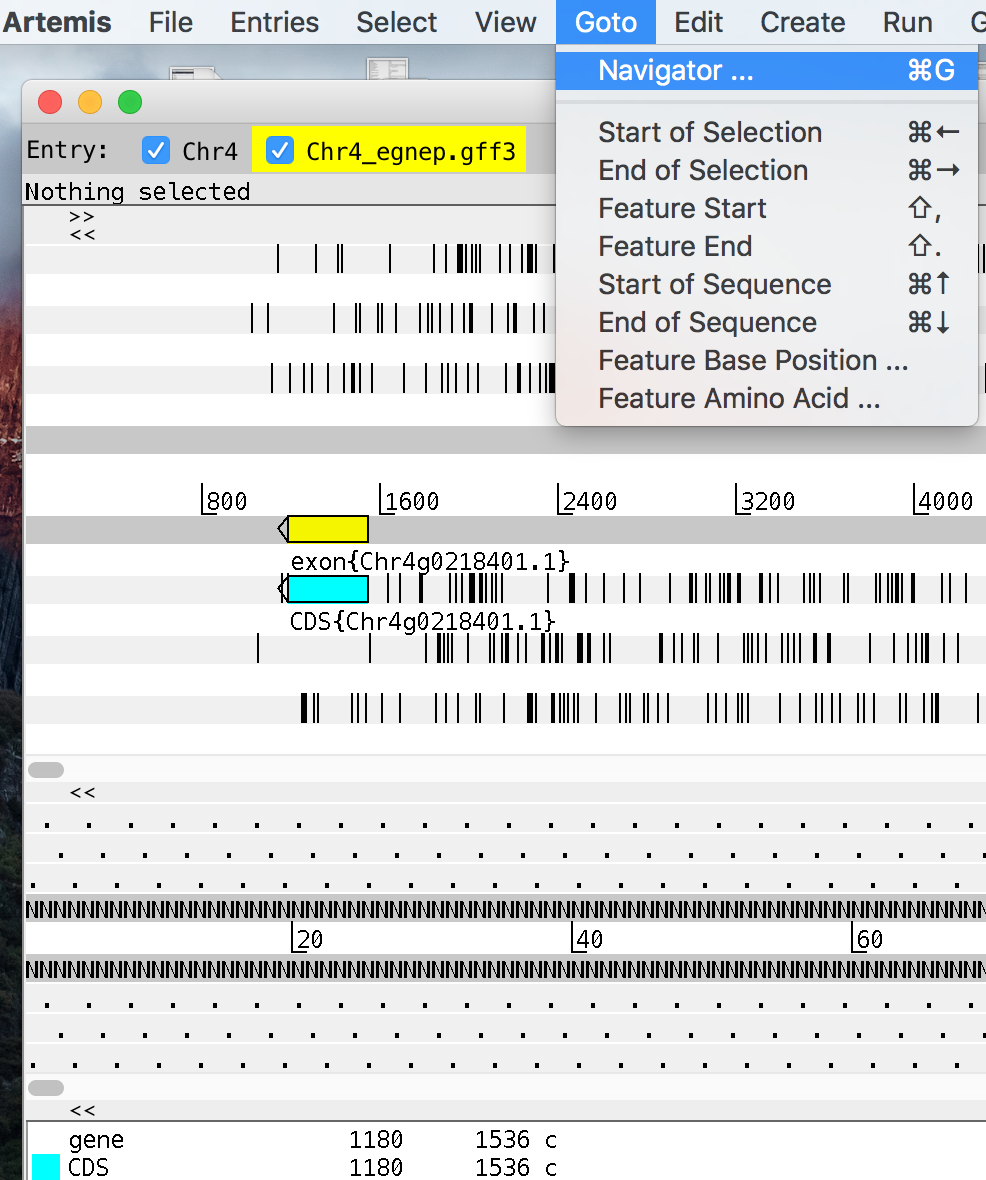
- Goto navigator

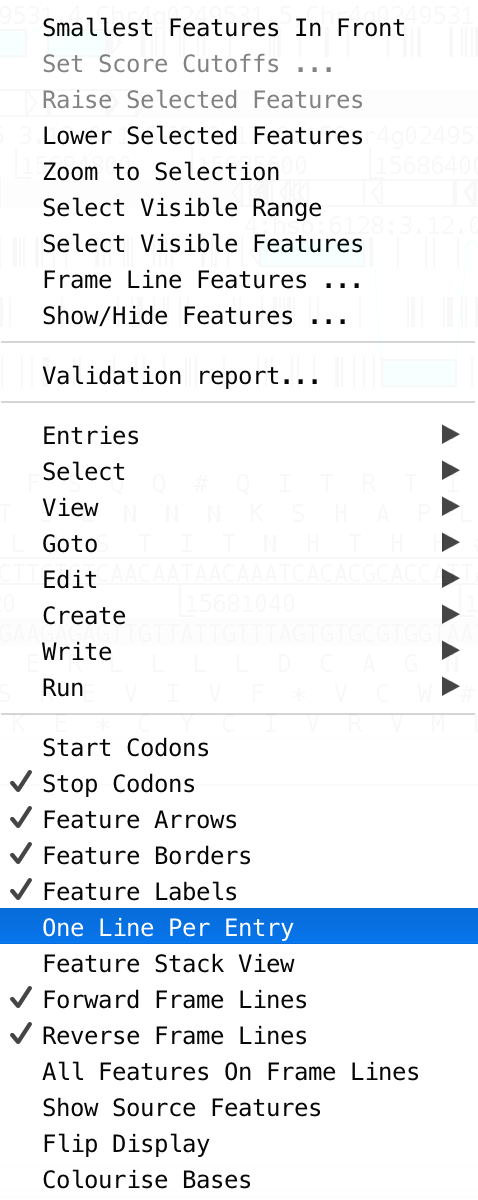
- To change the feature visusalisation mode Click right & tick ‘One line per feature’ & ‘all features on frame line’ options
-
Next methionine: click on a CDS in cyan and type the ‘cmd Y’ (Mac) or ‘ctrl Y’ (Windows / Linux)
- Undo: clicking on a CDS in cyan and type the ‘cmd Y’ (Mac) or ‘ctrl Y’ (Windows / Linux)
Annotate AT4G32500 :
- Shorter an exon and CDS & save
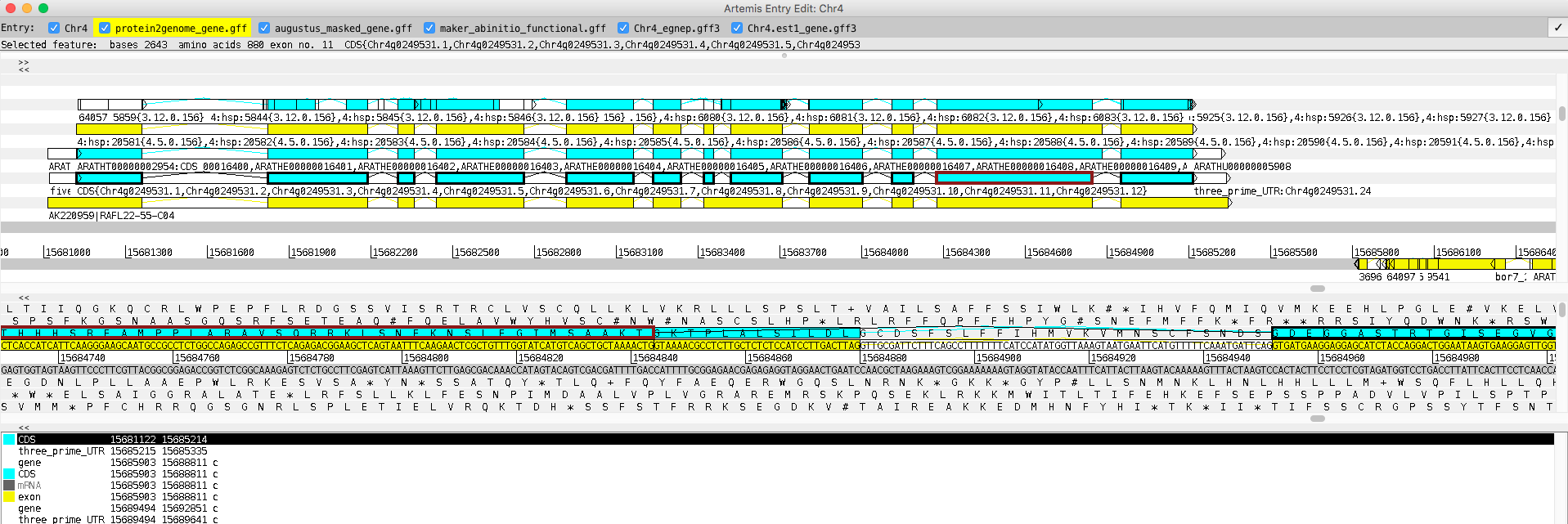 CDS(15684277..15684879) -> CDS(15684277..15684843)
CDS(15684277..15684879) -> CDS(15684277..15684843) - Check the functional annotation Open the Gene Builder by clicking on a feature and then typing the ‘cmd E’ (Mac) or ‘ctrl E’ (Windows / Linux)
Annotate AT4G32510 :
-
Set smaller gene as ‘obsolete’
-
Set the correct boundaries of the first transcript gene complement(15685825..15688811)
mRNA complement(15685903..15688811)
CDS complement(join(15685903..15686359,15686452..15686777,15686908..15686994,15687072..15687167,15687259..15687432,15687523..15687689,15687772..15687937,15688025..15688163,15688258..15688450,15688544..15688626,15688708..15688811))
- Duplicate the transcript, set the correct boundaries of the second alternative transcript and save
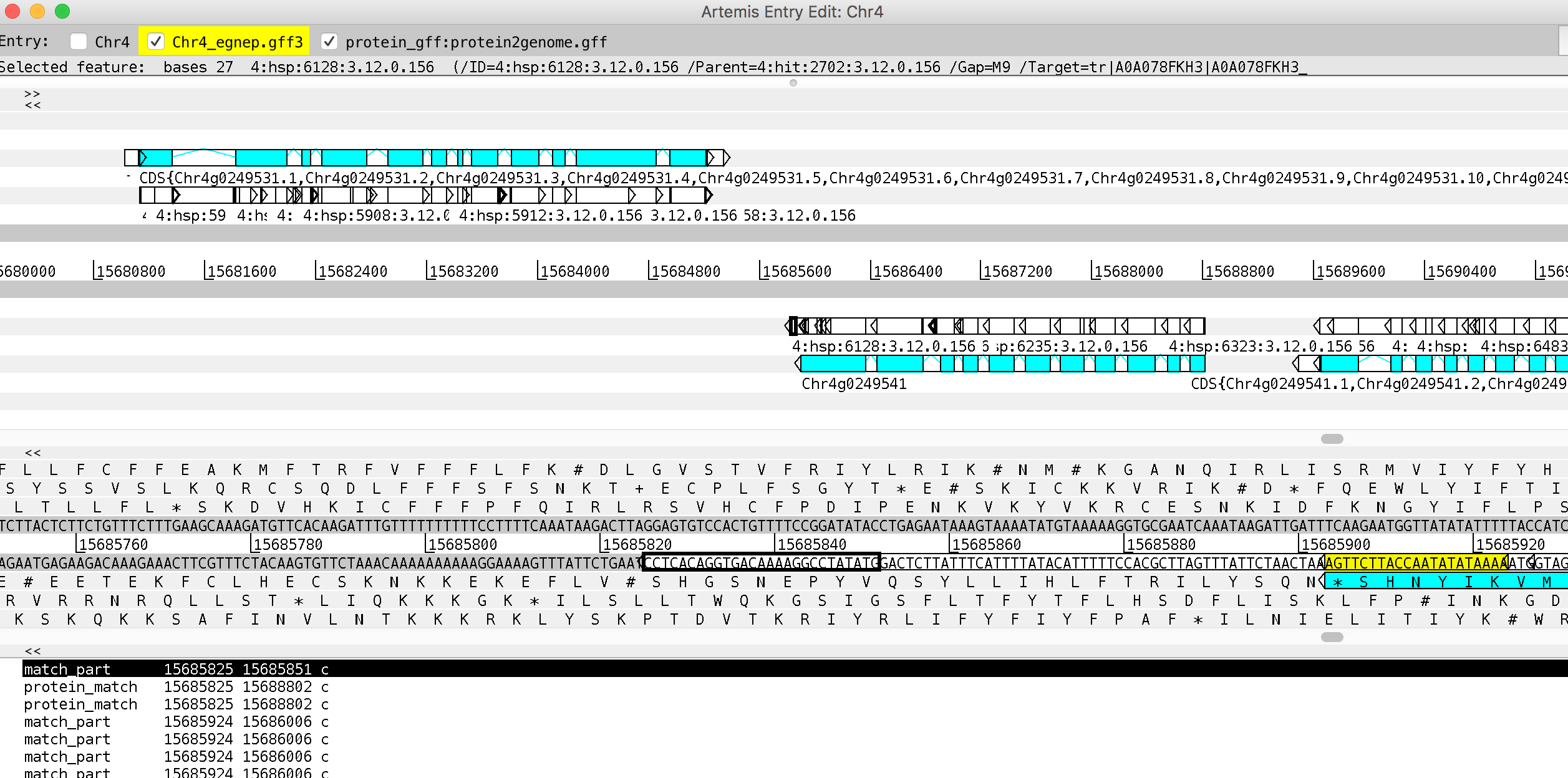 mRNA.1 complement(15685825..15688811)
CDS.1 complement(join(15685825..15685851,15685927..15686069,15686115..15686359,15686452..15686767,15686874..15686994,15687072..15687167,15687259..15687432,15687523..15687689,15687772..15687937,15688025..15688163,15688258..15688450,15688544..15688626,15688708..15688811))
mRNA.1 complement(15685825..15688811)
CDS.1 complement(join(15685825..15685851,15685927..15686069,15686115..15686359,15686452..15686767,15686874..15686994,15687072..15687167,15687259..15687432,15687523..15687689,15687772..15687937,15688025..15688163,15688258..15688450,15688544..15688626,15688708..15688811))
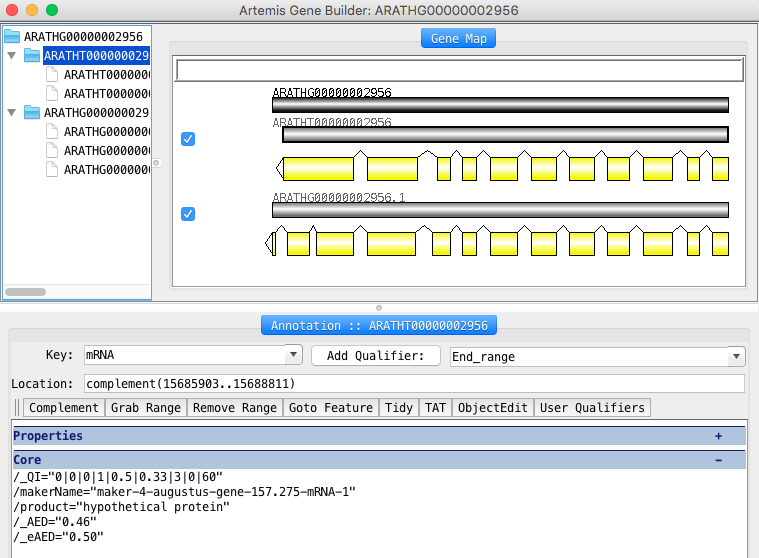
- Write polypeptide sequence, make blastp on uniprot KB to curate the functional annotation (product, gene_symbol) and save
